About Genome Comparator
Genome Comparator is used to compare whole genome data of isolates within a database using either the database defined loci (regions) or the coding sequences of an annotated genome as the comparator.
Here, we implement Genome Comparator to allow users to compare genomes across curated collections of genomic data hosted on our platform.
Why is it useful?
UNDER REVIEW ⚠️Genome Comparator helps researchers identify similarities and differences between genomes to understand genetic relationships, track evolutionary changes, and identify potential functional differences.
Some use cases include ...
How to use it?
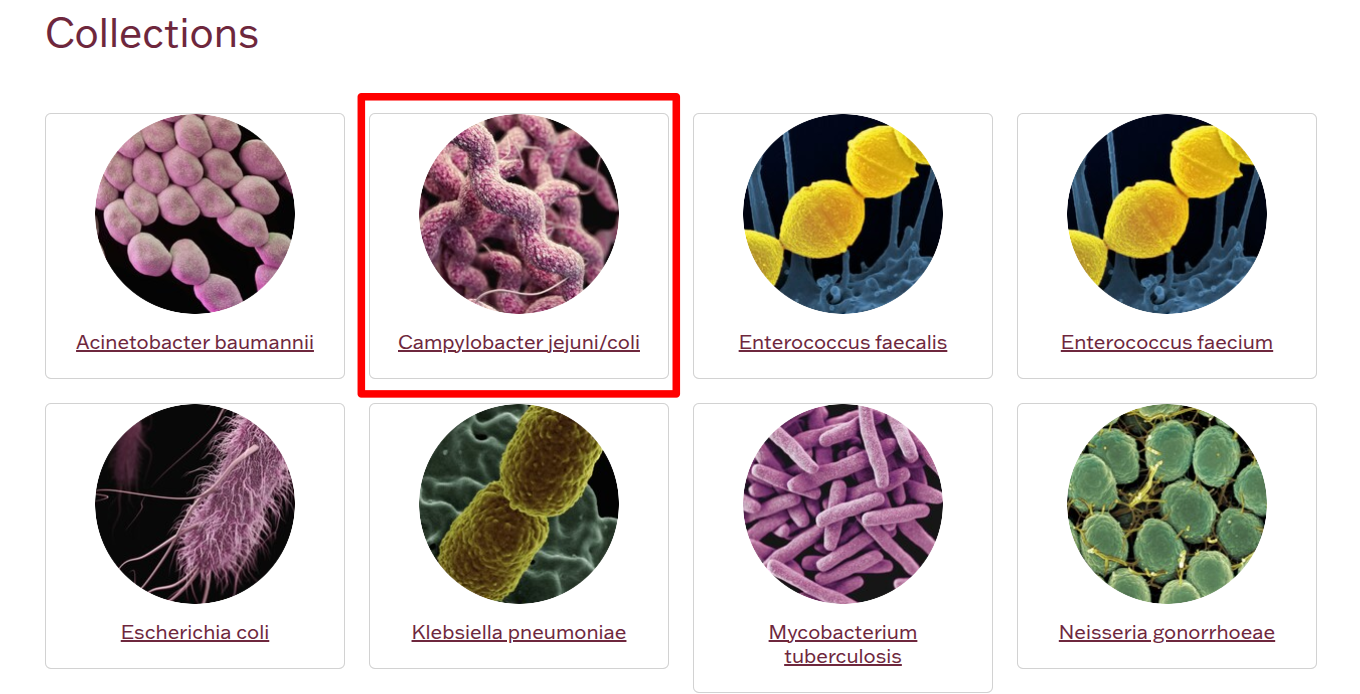
STEP 1 Go to the collections page to select the organism's database you want to search against.
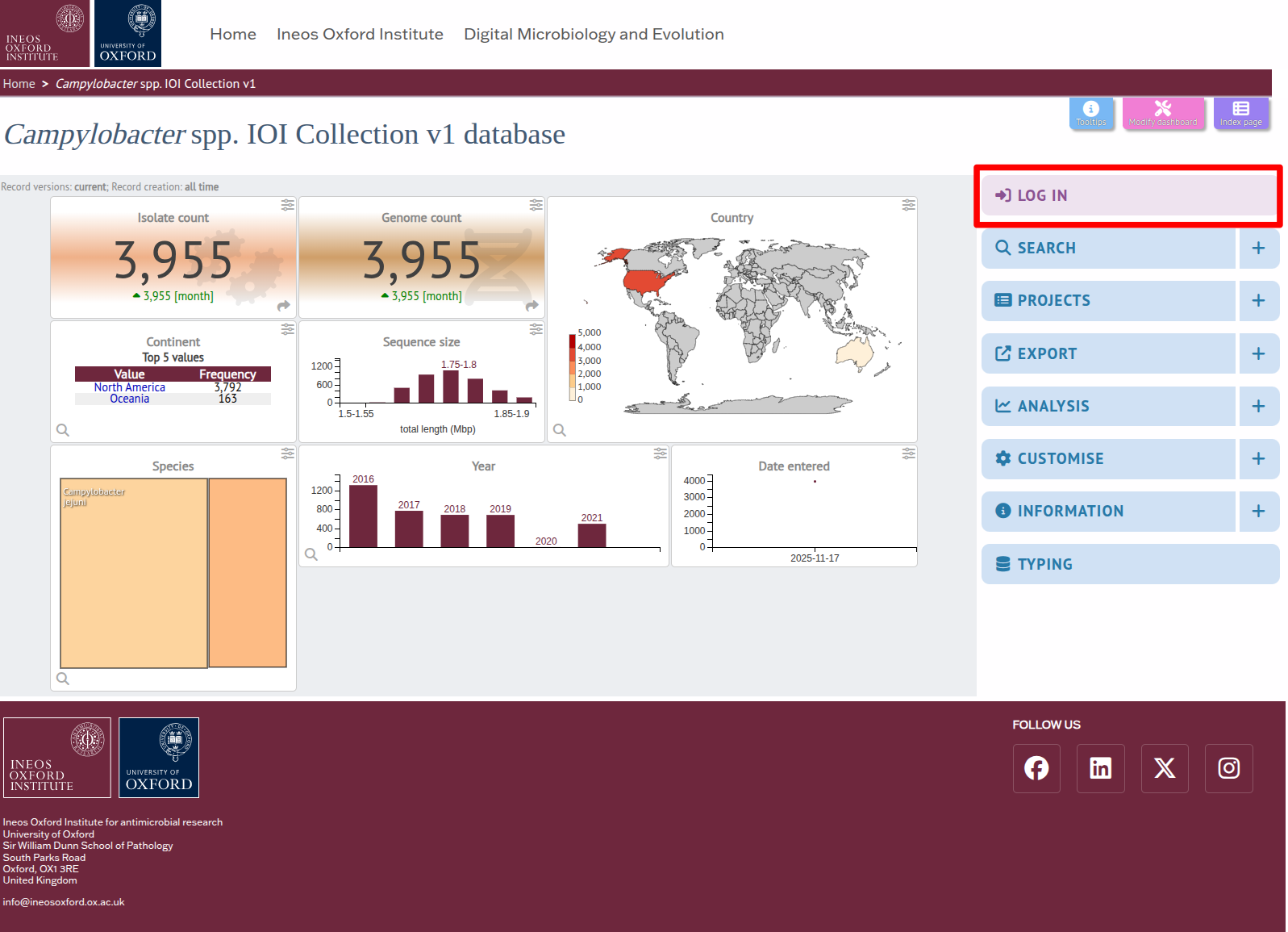
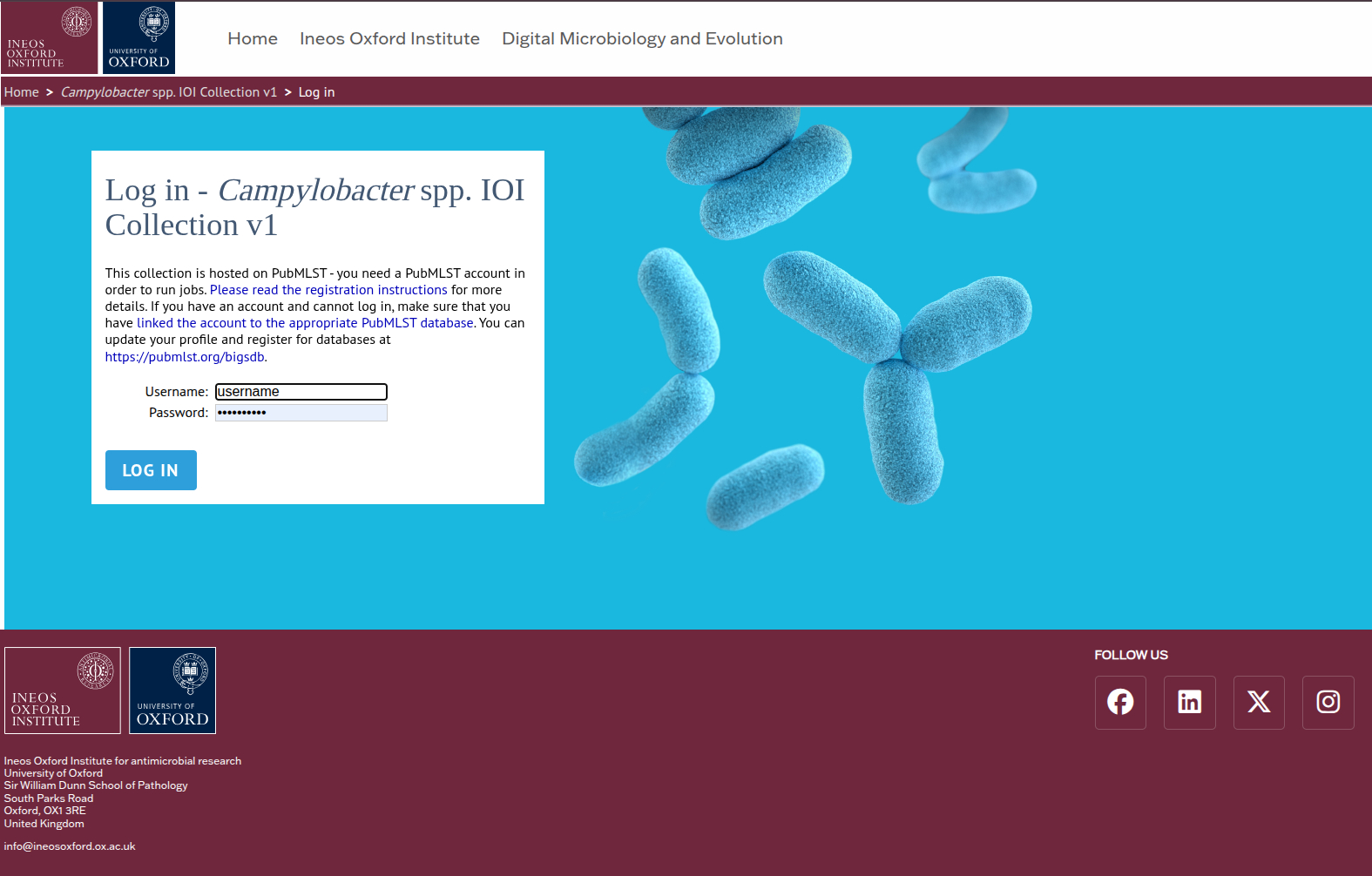
STEP 2 Log in using your PubMLST credentials.
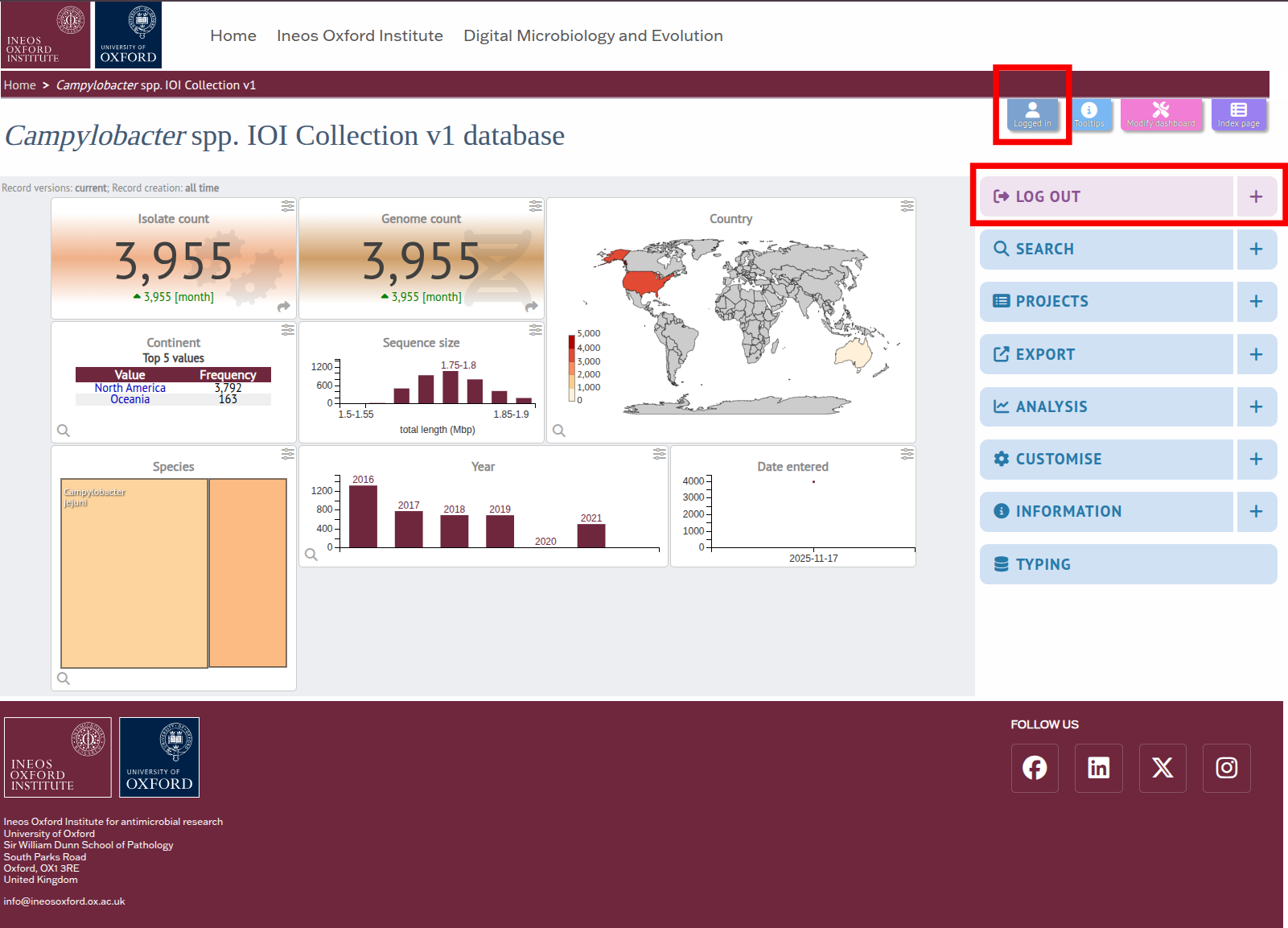
STEP 3 To access the tool's page, either click on the Analysis tab to display a link or find a button to the plugin at the bottom of a search results.
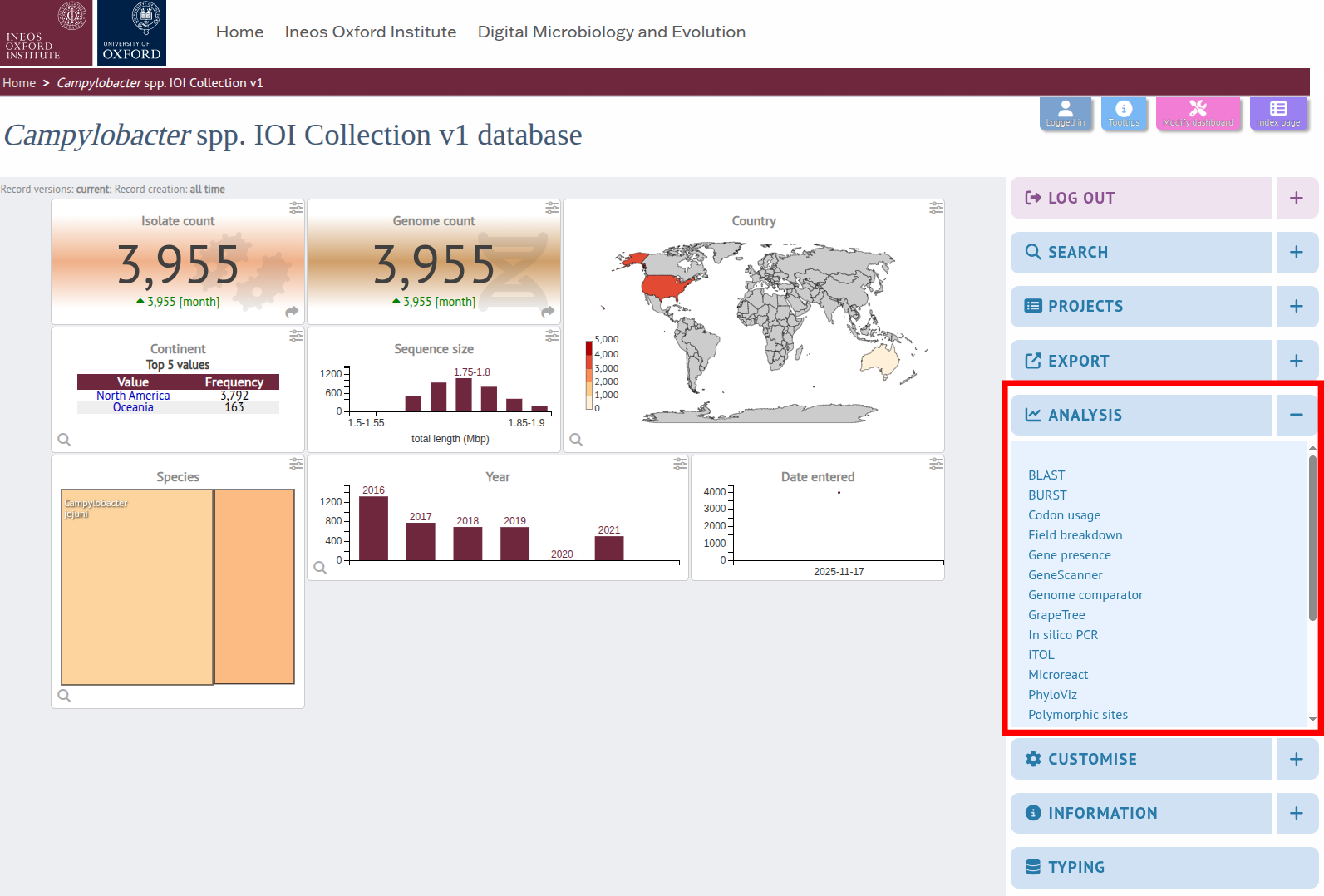
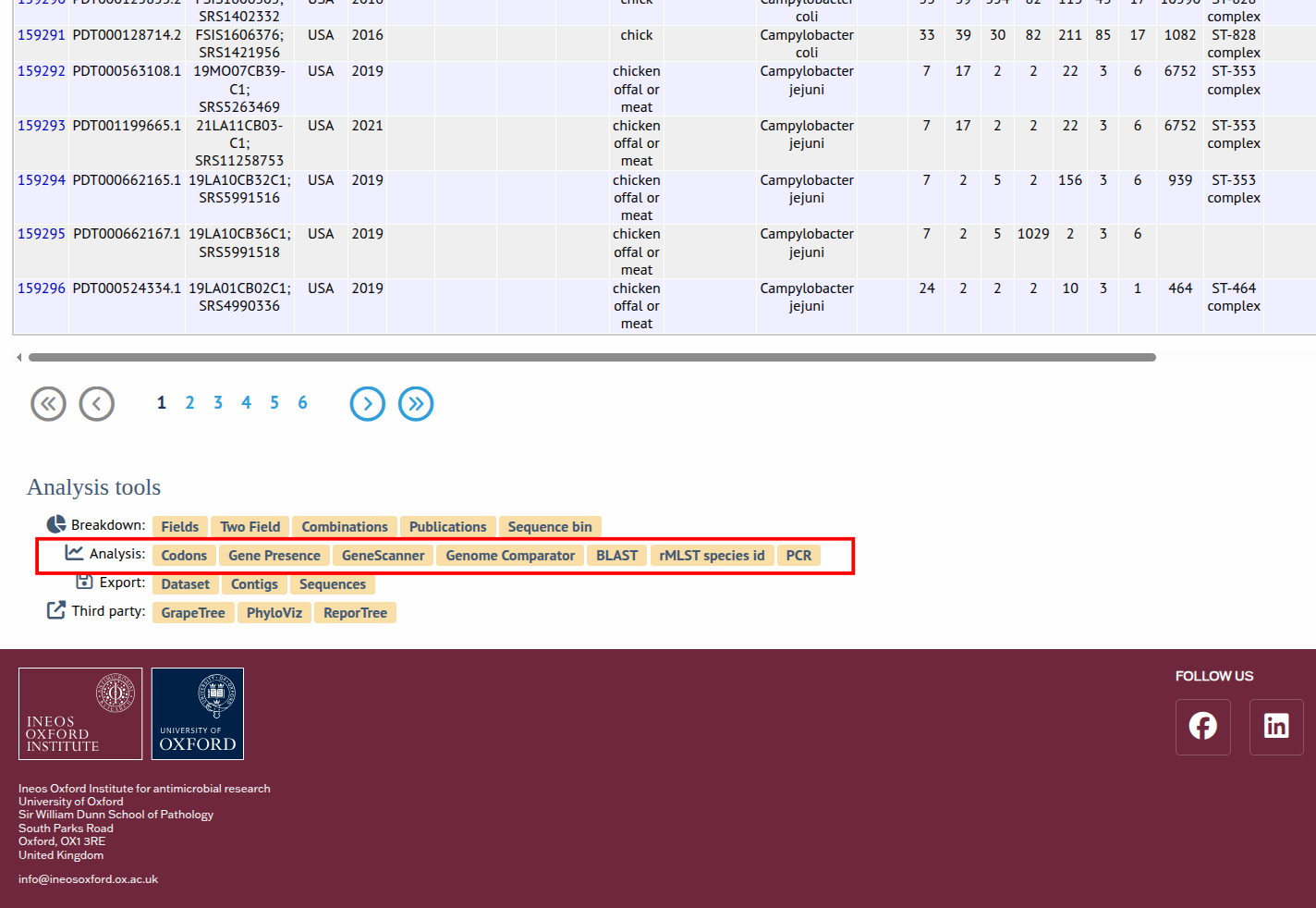
STEP 4 Once you see the Genome Comparator interface, choose the comparator scheme (database loci or annotated genome CDS), upload/select isolates, configure thresholds, and run the comparison.
UNDER REVIEW ⚠️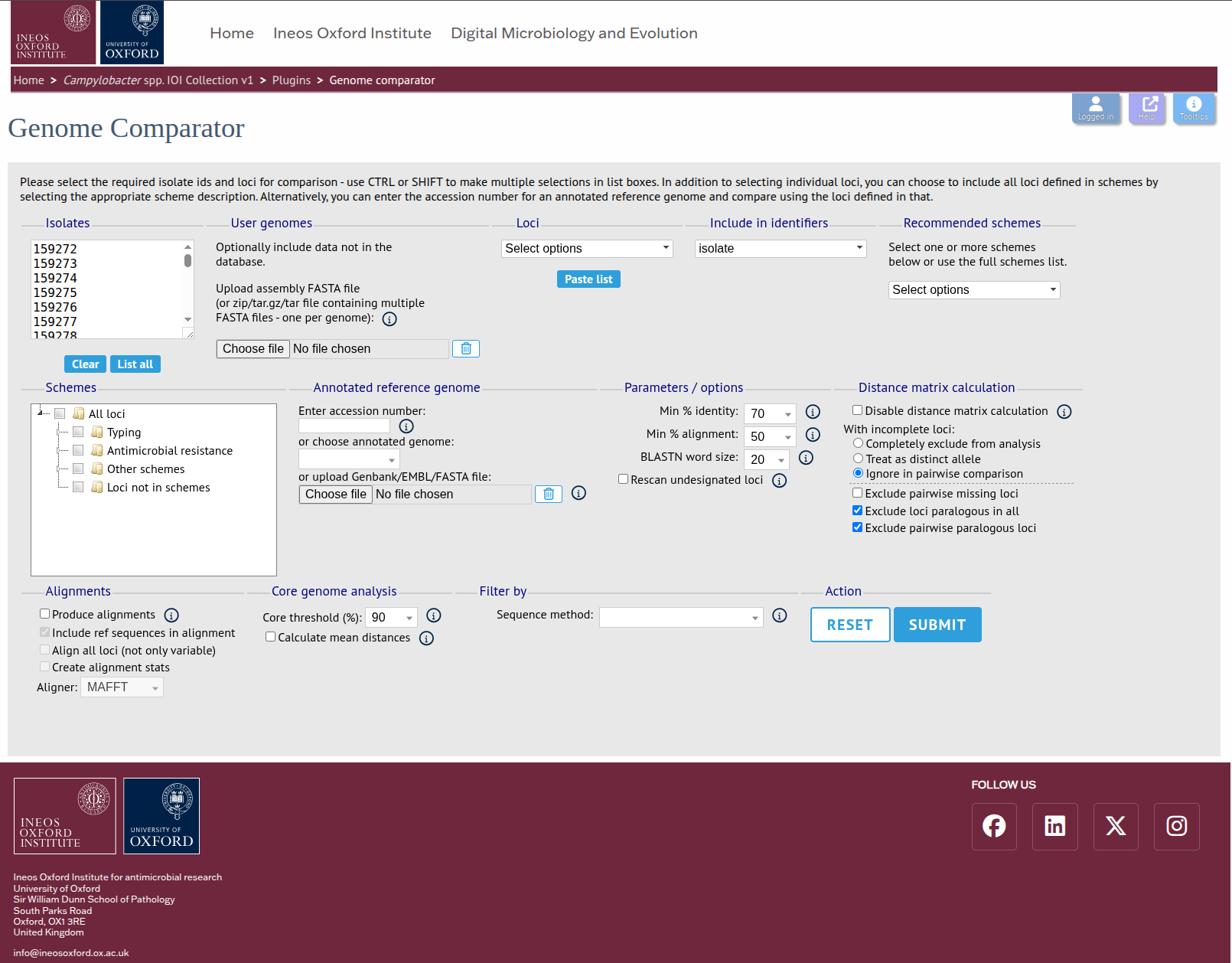
Contact us
For queries and to report any issues, email us at: email[at]biology.ox.ac.uk
